用R进行G×E互作的AMMI模型分析
在多地点试验中,突出的一个问题是林木基因型与环境之间往往存在显著的交互作用(Genotype by environment interaction, GEI),因此如何准确评估GEI对于后续林木良种的选育和推广至关重要。目前,农业上大多使用联合回归法1、主效可加互作可乘模型(Additive main effects multiplicative interaction, AMMI)2和基因型主效加基因型-环境互作效应双标图(Genotype Main Effect plus Genotype-by-Environment Interaction biplot, GGE双标图) 3分析多点试验,并据此来进行品种评价、试验点评价和品种生态区划分,其中GGE双标图越来越受关注。迄今,GGE双标图在林木上的应用4仍然很少,仅在少数树种如辐射松、杨树和乐昌含笑中有过报道。
00 示例数据集
示例采用agricolae包的数据集plrv。
|
|
01 使用AMMI()建模
使用agricolae包的AMMI()建模,具体代码如下:
|
|
现在可查看方差分析结果与GEI部分的主成分结果如下:
|
|
如果只保留前2个主成分,则可通过自编函数来操作:
|
|
|
|
查看ASV和YSI结果:
|
|
查看各基因型的PCs得分:
|
|
02 结果可视化
可以使用plot()和AMMI.contour()来生成双标图:
|
|
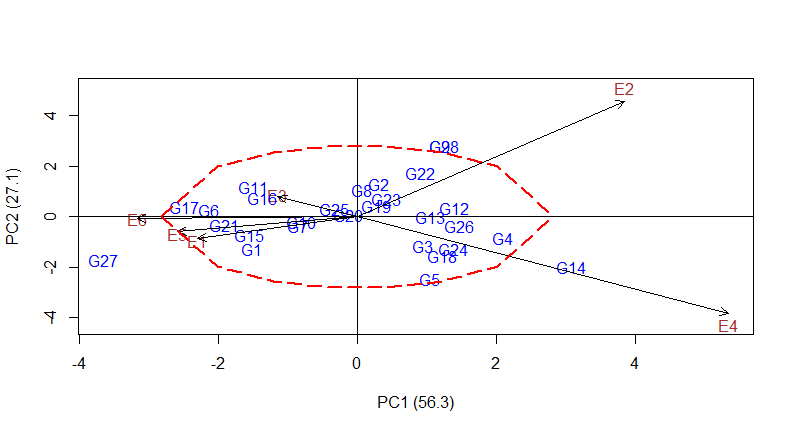
图1 AMMI模型的双标图
对于GEI的结果可视化,不如GGE方法来得直观。后一篇博文将介绍如何使用R来可视化GGE方法。
03 方法局限性
3.1 AMMI 模型的局限性
- 只限定于固定效应模型;
- 假定各地点误差同质;
- 要求数据是平衡的。
3.2 AMMI 模型可视化的局限性
- 未能充分展示基因型的稳定性和高产性;
- 未能充分展示试验地点间的关系。
04 参考文献
- Finlay K W, Wilkinson G N. The analysis of adaptation in a plant breeding programme [J]. Aust J Agric Res, 1963, 14: 742-754 [return]
- Gauch H G, Zobel R W. Identifying mega-environments and targeting genotypes [J]. Crop Sci, 1997, 37: 311-326 [return]
- Yan W. GGEbiplot—a Windows application for graphical analysis of multi-environment trial data and other types of two-way data [J]. Agron J, 2001, 93:1111-1118 [return]
- 程玲,张心菲, 张鑫鑫,等. 基于BLUP和GGE双标图的林木多地点试验分析[J]. 西北农林科技大学学报(自然科学版), 2018,46(3):87-93 [return]
文章标题:用R进行G×E互作的AMMI模型分析
文章作者:Yuanzhen Lin
创建时间:2018-08-14
原始链接:/programming/r/ammi-in-r/
